Full step-by-step example of fitting a GLM to experimental data and visualizing the results.
More specifically:








Script output:
Contrast 1 out of 21: left-right
Contrast 2 out of 21: audio
Contrast 3 out of 21: damier_V
Contrast 4 out of 21: V-H
Contrast 5 out of 21: clicGaudio
Contrast 6 out of 21: calculaudio
Contrast 7 out of 21: H-V
Contrast 8 out of 21: phrasevideo
Contrast 9 out of 21: video-audio
Contrast 10 out of 21: clicDaudio
Contrast 11 out of 21: clicDvideo
Contrast 12 out of 21: audio-video
Contrast 13 out of 21: phraseaudio
Contrast 14 out of 21: sentences
Contrast 15 out of 21: reading-visual
Contrast 16 out of 21: video
Contrast 17 out of 21: calculvideo
Contrast 18 out of 21: clicGvideo
Contrast 19 out of 21: computation-sentences
Contrast 20 out of 21: computation
Contrast 21 out of 21: damier_H
Python source code: plot_localizer_analysis.py
from os import mkdir, path
import numpy as np
import matplotlib.pyplot as plt
from nibabel import save
from nipy.modalities.fmri.glm import FMRILinearModel
from nipy.modalities.fmri.design_matrix import make_dmtx
from nipy.modalities.fmri.experimental_paradigm import \
load_paradigm_from_csv_file
from nipy.labs.viz import plot_map, cm
#######################################
# Data and analysis parameters
#######################################
# timing
n_scans = 128
tr = 2.4
# paradigm
frametimes = np.linspace(0.5 * tr, (n_scans - .5) * tr, n_scans)
# write directory
write_dir = 'results'
if not path.exists(write_dir):
mkdir(write_dir)
########################################
# Design matrix
########################################
paradigm = load_paradigm_from_csv_file('localizer_paradigm.csv')['0']
design_matrix = make_dmtx(frametimes, paradigm,
hrf_model='canonical with derivative',
drift_model="cosine", hfcut=128)
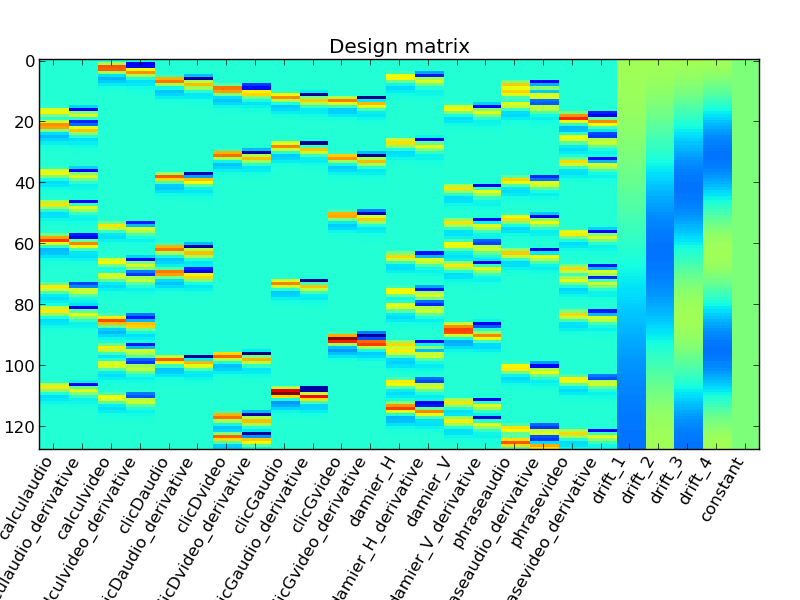
# Plot the design matrix
ax = design_matrix.show()
ax.set_position([.05, .25, .9, .65])
ax.set_title('Design matrix')
plt.savefig(path.join(write_dir, 'design_matrix.png'))
#########################################
# Specify the contrasts
#########################################
# simplest ones
contrasts = {}
n_columns = len(design_matrix.names)
for i in range(paradigm.n_conditions):
contrasts['%s' % design_matrix.names[2 * i]] = np.eye(n_columns)[2 * i]
# and more complex/ interesting ones
contrasts["audio"] = contrasts["clicDaudio"] + contrasts["clicGaudio"] +\
contrasts["calculaudio"] + contrasts["phraseaudio"]
contrasts["video"] = contrasts["clicDvideo"] + contrasts["clicGvideo"] + \
contrasts["calculvideo"] + contrasts["phrasevideo"]
contrasts["left-right"] = (contrasts["clicGaudio"] + contrasts["clicGvideo"]
- contrasts["clicDaudio"] - contrasts["clicDvideo"])
contrasts["computation"] = contrasts["calculaudio"] + contrasts["calculvideo"]
contrasts["sentences"] = contrasts["phraseaudio"] + contrasts["phrasevideo"]
contrasts["H-V"] = contrasts["damier_H"] - contrasts["damier_V"]
contrasts["V-H"] = contrasts["damier_V"] - contrasts["damier_H"]
contrasts["audio-video"] = contrasts["audio"] - contrasts["video"]
contrasts["video-audio"] = contrasts["video"] - contrasts["audio"]
contrasts["computation-sentences"] = contrasts["computation"] - \
contrasts["sentences"]
contrasts["reading-visual"] = contrasts["sentences"] * 2 - \
contrasts["damier_H"] - contrasts["damier_V"]
########################################
# Perform a GLM analysis
########################################
fmri_glm = FMRILinearModel('s12069_swaloc1_corr.nii.gz',
design_matrix.matrix, mask='compute')
fmri_glm.fit(do_scaling=True, model='ar1')
#########################################
# Estimate the contrasts
#########################################
for index, (contrast_id, contrast_val) in enumerate(contrasts.items()):
print(' Contrast % 2i out of %i: %s' %
(index + 1, len(contrasts), contrast_id))
# save the z_image
image_path = path.join(write_dir, '%s_z_map.nii' % contrast_id)
z_map, = fmri_glm.contrast(contrast_val, con_id=contrast_id, output_z=True)
save(z_map, image_path)
# Create snapshots of the contrasts
vmax = max(-z_map.get_data().min(), z_map.get_data().max())
plot_map(z_map.get_data(), z_map.get_affine(),
cmap=cm.cold_hot, vmin=-vmax, vmax=vmax,
slicer='z', black_bg=True, threshold=2.5,
title=contrast_id)
plt.savefig(path.join(write_dir, '%s_z_map.png' % contrast_id))
plt.show()
Total running time of the example: 128.34 seconds ( 2 minutes 8.34 seconds)